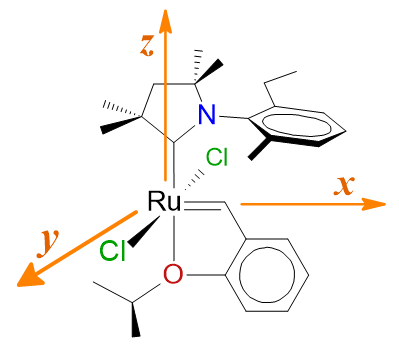
Catalysts are also represented by means of SMILES strings. For example, the first entry in Marx2015 (or entry 487 in the database, CAAC.09):
To facilitate inspections, we define the Cartesian positive directions as illustrated in the figure. Accordingly, the SMILES string begins with the ligands positioned along the x-axis, typically representing the Ru=C carbene bond. In this case, the ligand is a 2-isopropoxy-benzylidene group:

Next, the ligands occupying the ±y directions, usually characterized as anionic ligands:
Finally, the ligands positioned along the z-axis, often corresponding to NHC (N-heterocyclic carbene) and CAAC (cyclic alkyl amino carbene) ligands, are represented in the database:
It should be noted that there is no ligand in the -x direction. However, the -z direction is left unspecified to prevent repetition of the 2-isopropoxy-benzylidene ligand, which is assigned to the +x direction.
The SMILES strings stored under the Catalyst field of the CatalySeed database are not directly compatible with the RDKit workflow. However, they share a consistent structural pattern—ylidene-[Ru]-anions followed by stabilizing ligands (e.g., phosphines, pyridines, NHCs, CAACs)—which facilitates their curation and conversion into catalytically relevant structures using standard text editors. Accordingly, the CatalySeed database also includes fields for PreCat (precatalyst) and MCB (metallocyclobutane) structures, which contain SMILES strings already formatted for RDKit-based conformer sampling. For instance, the SMILES for CAAC.09 are reported as follows:
| Catalyst | CC(C)Oc1ccccc1C=[Ru](Cl)(Cl)=C2C(CC(C)(C)N2c3c(CC)cccc3C)(C)(C) |
|---|---|
| PreCat | CC(C)[O+]7c1ccccc1[CH][Ru-]7(Cl)(Cl)[C]2C(CC(C)(C)N2c3c(CC)cccc3C)(C)(C) |
| MCB | CC(C)Oc1ccccc1[CH]7C[CH2][Ru]7(Cl)(Cl)[C]2C(CC(C)(C)N2c3c(CC)cccc3C)(C)(C) |